Show code cell source
%config InlineBackend.rc = {"figure.dpi": 72, "figure.figsize": (6.0, 4.0)}
%matplotlib inline
import abtem
import ase
import matplotlib.pyplot as plt
from ase.io import write
4D-STEM quickstart#
abtem.config.set({"device": "cpu"})
<abtem.core.config.set at 0x21a4e10ff40>
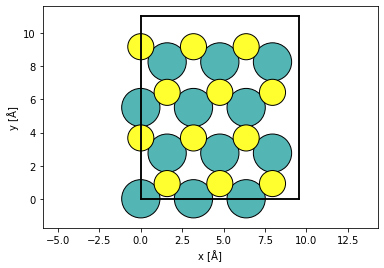
atoms = ase.build.mx2(vacuum=2)
atoms = abtem.orthogonalize_cell(atoms)
atoms *= (3, 2, 1)
abtem.show_atoms(atoms)
(<Figure size 432x288 with 1 Axes>,
<AxesSubplot: xlabel='x [Å]', ylabel='y [Å]'>)
potential = abtem.Potential(atoms, sampling=0.05)
probe = abtem.Probe(energy=80e3, semiangle_cutoff=30)
probe.grid.match(potential)
grid_scan = abtem.GridScan.from_fractional_coordinates(
potential,
start=[0, 0],
end=[1 / 3, 1 / 2],
sampling=probe.aperture.nyquist_sampling,
)
detector = abtem.PixelatedDetector()
measurements = probe.scan(potential, scan=grid_scan, detectors=detector)
measurements.array
|
||||||||||||||||
measurements.compute()
[########################################] | 100% Completed | 5.23 sms
<abtem.measurements.DiffractionPatterns object at 0x0000021A57CD9AB0>
filtered_measurements = measurements.gaussian_source_size(.3)
center_of_mass = filtered_measurements.center_of_mass()
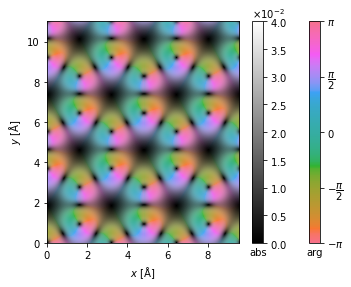
interpolated_center_of_mass = center_of_mass.interpolate(0.05).tile((3, 2))
interpolated_center_of_mass.show(cbar=True, vmax=0.04, vmin=0);