Show code cell source
%config InlineBackend.rc = {"figure.dpi": 72, "figure.figsize": (6.0, 4.0)}
%matplotlib inline
import matplotlib.pyplot as plt
from ase.build import graphene
from ase.io import write
import abtem
STEM quickstart#
This notebook demonstrates a basic simulation of a scanning transmission electron microscopy image of graphene with a silicon dopant.
Configuration#
We start by (optionally) setting our configuration. See documentation for details.
abtem.config.set({"device": "cpu", "fft": "fftw"})
<abtem.core.config.set at 0x249659ff760>
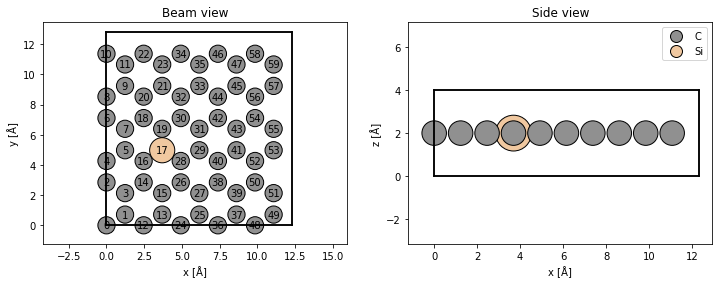
Atomic model#
We create an orthogonal graphene cell. See our walkthough or our tutorial on atomic models.
atoms = abtem.orthogonalize_cell(graphene(vacuum=2))
atoms *= (5, 3, 1)
We dope the graphene with silicon by changing the atomic number of one of the atoms.
atoms.numbers[17] = 14
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 4))
abtem.show_atoms(atoms, ax=ax1, title="Beam view", numbering=True, merge=False)
abtem.show_atoms(atoms, ax=ax2, plane="xz", title="Side view", legend=True);
Potential#
We create an ensemble of potentials using the frozen phonon model. See our walkthrough on frozen phonons.
frozen_phonons = abtem.FrozenPhonons(atoms, 8, sigmas=0.1)
We create a potential from the frozen phonons model, see walkthrough on potentials.
potential = abtem.Potential(frozen_phonons, sampling=0.05)
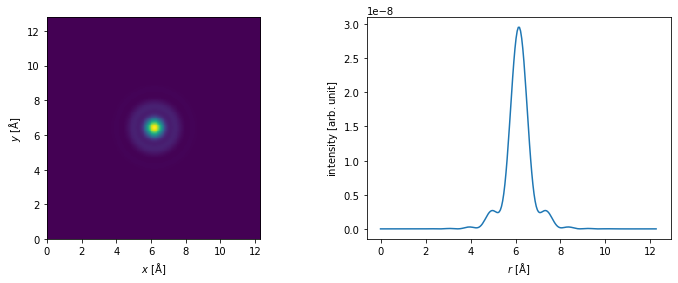
Wave function#
We create a probe wave function at an energy of \(80 \ \mathrm{keV}\), an objective aperture of \(30 \ \mathrm{mrad}\), a spherical aberration of \(10 \ \mu\mathrm{m}\) and Scherzer defocus. See our walkthrough on wave functions.
Partial temporal coherence is neglected here, see our tutorial on partial coherence.
probe = abtem.Probe(energy=80e3, semiangle_cutoff=25, Cs=10e4, defocus="scherzer")
probe.grid.match(potential)
print(f"defocus = {probe.aberrations.defocus} Å")
print(f"FWHM = {probe.profiles().width().compute()} Å")
defocus = 79.14274499114904 Å
FWHM = 0.8963562846183777 Å
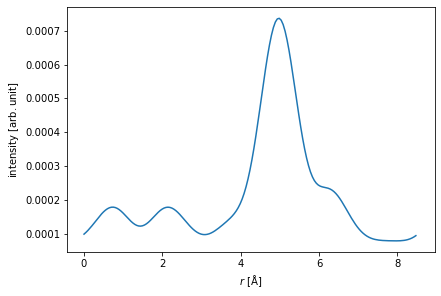
We show the profile of the probe.
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 4))
probe.show(ax=ax1)
probe.profiles().show(ax=ax2);
[########################################] | 100% Completed | 111.71 ms
[########################################] | 100% Completed | 115.02 ms
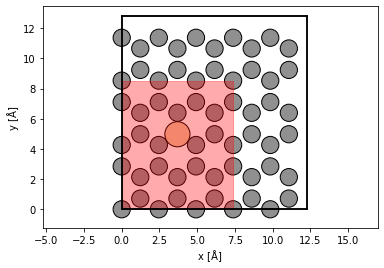
Scan#
We select a scan region using fractional coordinates. We scan at the Nyquist frequency, allowing us to interpolate the measurements below.
grid_scan = abtem.GridScan(
start=(0, 0),
end=(3 / 5, 2 / 3),
sampling=probe.aperture.nyquist_sampling,
fractional=True,
potential=potential,
)
fig, ax = abtem.show_atoms(atoms)
grid_scan.add_to_plot(ax)
Scan and detect#
We use a flexible annular detector, this will let us choose the detector angles after the running multislice.
detector = abtem.FlexibleAnnularDetector()
We run the scanned multislice algorithm. See our walkthrough on scanning and detecting.
flexible_measurement = probe.scan(potential, scan=grid_scan, detectors=detector)
flexible_measurement.compute()
[########################################] | 100% Completed | 11.01 ss
<abtem.measurements.PolarMeasurements object at 0x000002496C904580>
Integrate measurements#
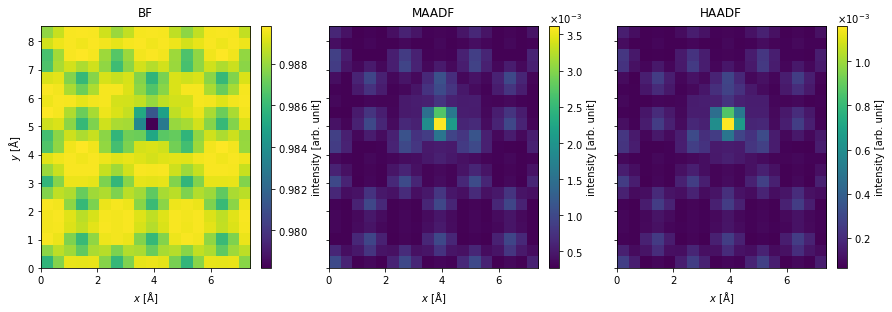
The measurements are integrated to obtain the bright field, medium-angle annular dark field and high-angle annular dark field signals.
bf_measurement = flexible_measurement.integrate_radial(0, probe.semiangle_cutoff)
maadf_measurement = flexible_measurement.integrate_radial(50, 150)
haadf_measurement = flexible_measurement.integrate_radial(90, 200)
The measurements are stacked and shown as an exploded plot.
measurements = abtem.stack(
[bf_measurement, maadf_measurement, haadf_measurement], ("BF", "MAADF", "HAADF")
)
measurements.show(
explode=True,
figsize=(14, 5),
cbar=True,
);
Postprocessing#
Typically some post-processing steps are necessary to obtain the final results, see our walkthrough on scanning and detecting for more details
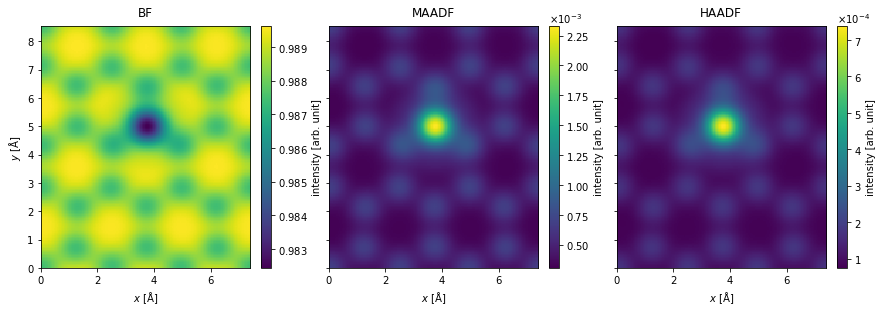
The measurements are interpolated to a sampling rate of \(0.05 \ \mathrm{Å / pixel}\).
interpolated_measurements = measurements.interpolate(0.05)
We can simulate partial spatial coherence by applying a gaussian filter. The standard deviation of the filter is \(0.3 \ \mathrm{Å}\), the approximate size of the electron source.
filtered_measurements = interpolated_measurements.gaussian_filter(0.3)
The interpolated and filtered measurements shown as an exploded plot.
filtered_measurements.show(
explode=True,
figsize=(14, 5),
cbar=True,
);
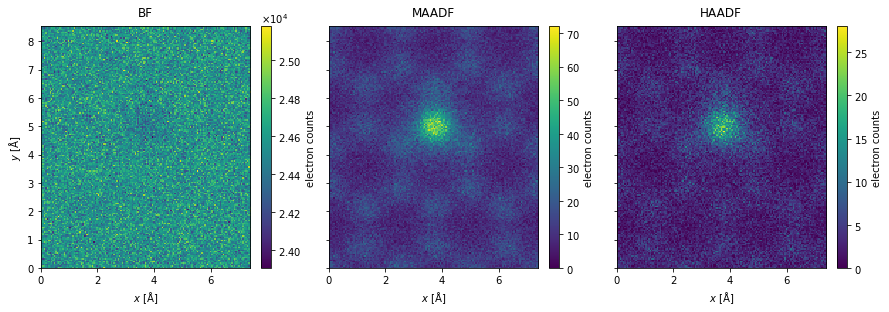
We simulate a finite electron dose of \(10^7 \ \mathrm{e}^- / \mathrm{Å}^2\) by applying poisson noise
noisy_measurements = filtered_measurements.poisson_noise(dose_per_area=1e7)
noisy_measurements.show(
explode=True,
figsize=(14, 5),
cbar=True,
);
Showing the results as a line profile often provides a better sense of relative intensities.
line_profile = filtered_measurements.interpolate_line(
start=(1 / 2, 0), end=(1 / 2, 1), fractional=True
)
line_profile[-1].show();