Show code cell source
%config InlineBackend.rc = {"figure.dpi": 72, "figure.figsize": (6.0, 4.0)}
%matplotlib inline
import ase
import matplotlib.pyplot as plt
from abtem.tilt import precession_tilts
from ase.io import read
import abtem
PED quickstart#
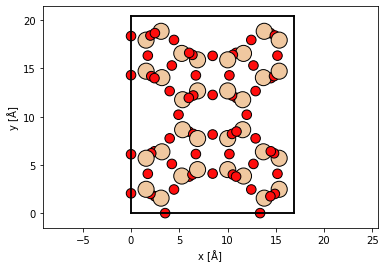
atoms = read("data/SiO2_zeolite.cif")
cell = atoms.cell.copy()
atoms *= (1, 1, 20)
abtem.show_atoms(atoms, plane="xy");
frozen_phonons = abtem.FrozenPhonons(atoms, 4, sigmas=0.078)
potential = abtem.Potential(
frozen_phonons,
sampling=.5,
projection="infinite",
slice_thickness=1,
)
wave = abtem.PlaneWave(energy=100e3)
wave.grid.match(potential)
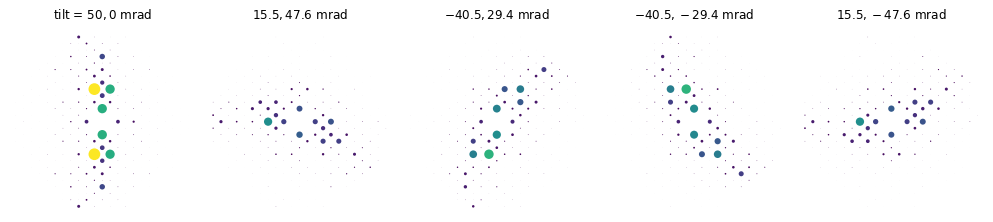
wave.tilt = precession_tilts(50, 5)
measurement = wave.multislice(potential).diffraction_patterns().mean(0).compute()
[########################################] | 100% Completed | 2.46 sms
spots = (
measurement.crop(25)
.block_direct()
.index_diffraction_spots(cell=atoms, threshold=1e-5)
)
visualization = spots.show(explode=True, scale=0.8, figsize=(16, 5))
visualization.axis_off(spines=False)